The Body Scientific, July 28, 2023
Gene Mining for New Antibiotics
Nineteen Chemists and Biologists From Jena
Antibiotics To Help Gardens and Forests
Part I
Richard Kessin
The title of a recent paper in the Journal of the American Chemical Society, or JACS for short, is: Ecological Niche-Inspired Genome Mining Leads to the Discovery of Crop-Protecting Non-ribosomal Lipopeptides Featuring a Transient Amino Acid Building Block. JACS publishes heavy stuff. Unreadable perhaps, but serious. Readers may think these authors and JACS are on a one lane road to obscurity, but there is substance here and a huge amount of work. A shorter title might have been Antibiotics from Pseudomonas Bacteria Kill Nasty Amoebae and Fungi. Academic paper titles are sometimes like the sausage Congress makes of legislation. The authors named the new class of antibiotics after Keanu Reeves, the Canadian actor who plays a retired assassin, John Wick, and who emerges to kill bad guys. There are many diseases caused by amoebae and fungi and they are all bad guys. Forestry, agriculture, and medicine have few defenses against these organisms. Fungi kill our crops and trees in periodic waves. We depend on the forest , and I want our ash, chestnut, hemlock, and elm trees back, or at least to give them a fighting chance. These compounds could help.
The Jena people, led by Dr. Pierre Stallforth with whom I spoke recently, have a strategy that lets evolution do much of the work. They looked for antibiotics in biological situations where two or more species have fallen into an equilibrium, a condition biologists call mutualism, or competition. They reasoned that one species may make an antibiotic or other natural product to compete. There is probably no limit to the number of niches with competing species in biology.
First, a word about amoebae, or amoeba-like cells, the predators of bacteria. These cells have a lot of internal architecture: vacuoles, nuclei, sites to make special proteins, structures to carry out tasks of digestion and energy production, not to mention ways to recognize harmful bacteria and pull them inside. Amoebae-like cells crawl over the surface of our lungs, peritoneum, and kidneys. All higher organisms have amoebae or similar cells as part of their cellular repertoire. Think of them as a general and essential cellular life form that evolution has kept. Specific species of amoebae can also cause disease; think of amoebic dysentery or the ghastly brain destroying amoebae swimmers get from freshwater ponds.
Nasty bacteria have defenses against predator amoebae. Some take advantage of being engulfed and live in a special vacuole, surrounded by a membrane where they have adapted to interrupt a process that normally kills them. They hide in plain sight, but it takes effort. Mycobacterium tuberculosis and Legionella pneumoniae live in cellular compartments, where they grow and divide. The Stallforth lab uses a species called Dictyostelium discoideum that lives in soil and eats the many kinds of bacteria they find there. Legionella and TB bacteria live happily in Dictyostelium amoebae.
Your columnist and his lab studied Dictyostelium for decades and wrote a book on these shapeshifting, complicated, and quite beautiful creatures. For a video of what they do, type ‘John Bonner and Princeton’ into a browser and you will find a lovely film, made in 1947. Albert Einstein asked to see it, so John walked to his house and projected the film on a bedsheet. There is, of course, more to this story, but let’s return to Dr. Stallworth.
Instead of isolating bacteria or amoebae from this niche and asking if individual cells produced antibiotics, they extracted DNA, which takes a few minutes, and examined the sequences of A, C, G, and T of millions of individual genes, which takes longer. It sounds hard, and it was once, but now the process is efficient and automated. Two classes of genes make the enzymes to produce antibiotics, each easily recognized by their DNA sequences stored in enormous databases. As an example, imagine reading a book and you come across the sentence Call me Ishmael. You would know that the book is Moby Dick. Never mind that it’s the first sentence. DNA is the same. Find a sequence that resembles previously studied antibiotic genes and there probably is an organism in your niche that makes an antibiotic. Then you can try to grow that organism, in this case Pseudomonas, and recover the antibiotic.
The story of our niche now takes us to the experimental forest of the University of Virginia in the Great Smoky Mountains in 2014, where evolutionary biologists Joan Strassmann and David Quellar of Washington University were looking for new strains of Dictyostelium. Joan found a fruiting body of Dictyostelium, which looks like a lollipop about 1mm high on a steaming pile of deer scat. Where the business end of the lollipop would be, there was a ball of tough spores, about 50,000 held together in a drop by surface tension. Spores are tough and survive desiccation and starvation. Fruiting bodies form in the lab, but that was the first time anyone had seen one in the wild. In the liquid around the spores, they later found, there were bacteria, a strain of Pseudomonas, now called QS1027, that secreted antibiotics. The Jena lab has now determined the chemical structures of these Keanumycins.
The image I would like to leave you with is Professor Joan Strassmann, author of an excellent recent book called Slow Birding, a member of The National Academic of Sciences, an exceptional teacher and mentor, on hands and knees with her nose six inches from a heap of deer poop, yelling in delight. I was on that field trip, and I looked too. Joy is where you find it.
This story is just beginning. The next column will explain more about the Keanomycins, what they kill, and how they work.
Prof. Pierre Stallforth is in the Department of Paleobiotechnology, Leibniz Institute for Natural Products Research and Infection Biology, Hans Knoll Institute, 07745, Jena, Germany, and the Faculty of Chemistry and Earth Sciences, Institute of Organic Chemistry and Macromolecular Chemistry, Friedrich Schiller University, Jena.
Richard Kessin, Ph.D is Emeritus Professor of Pathology and Cell Biology in the Department of Pathology and Cell Biology, College of Physicians and Surgeons, Columbia University Irving Medical Center. He has been writing the Body Scientific column since 2010. Earlier columns are at RichardKessin.com.
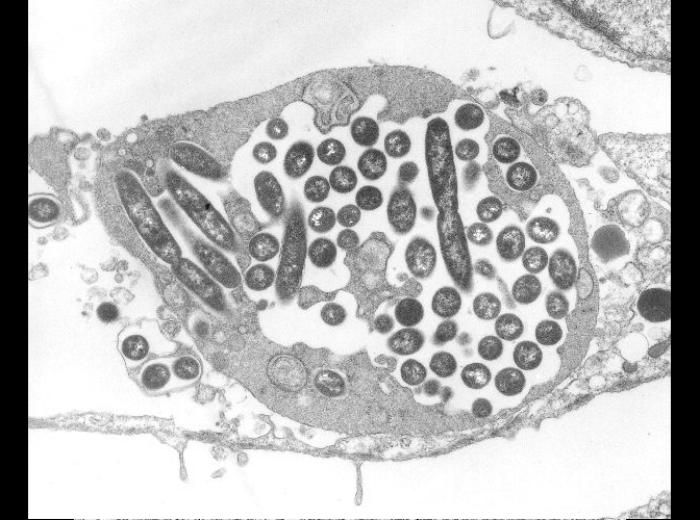
Legionella pneumophila growing in a vacuole of a human cell.
The Body Scientific, July 2023
Big Things, Momma, From Small Things Someday Come
Antibiotics and The Nitty Gritty Dirt Band
Part II
Richard Kessin
In the last column we met scientists Joan Strassmann and Pierre Stallforth, and left Joan, lying on the ground in a Virginia forest, peering through a magnifying glass at a pile of steaming deer scat. She saw the first Dictyostelium fruiting body in the wild, sprouting out of a pellet of poop. We thought that was fun, but a small thing, a curiosity. But something bigger came of it. (The Dirt Band are at Mashantucket on August 5). That song could be an anthem of science; progress usually springs from small starts. The origins of microbiology and much of medicine derive from Louis Pasteur’s experiments with crystals of sodium tartrate, followed by step including the germ theory of disease, that built over time.
In the 1960’s some physicians thought they had infection on the run, but they had not reckoned with the uncanny ability of bacteria to mutate to drug resistance. In 1961, I asked a pediatrician friend of my family if he could help me find antibiotics among the molds of Laconia, New Hampshire. Great idea! he said, and we set to work; I planned to exhibit at the high school science fair. I isolated molds and Dr. Baker taught me to spread bacteria on blood agar petri dishes and then put molds next to them. We hoped that secreted fungal products would kill renal E. coli and pathogenic Streptococcus bacteria. None did.
New antibiotics are still a priority, and some of the methods are the same as when Alexander Fleming discovered penicillin in the 1920’s. Find something that grows on a Petri dish and test it against bacteria. This approach has limitations–most microorganisms do not grow on Petri dishes filled with nutritious agar. Sebastian Götze and his colleagues in Pierre Stallforth’s lab in Germany developed a method to find antibiotic producing genes that avoids these problems. They call it “ecological niche genome mining”.
They found a group of organisms growing together in what ecologists and evolutionary biologists call a niche. The organisms compete but have produced a stable co-existence that could require production of an antibiotic by one or the other member of the community. The goal is to find the gene that produces that antibiotic. They do not have to grow the organisms on Petri dishes. At this point, scientists (or students) collect the community of cells and dissolve them in a detergent that destroys most molecules but leaves DNA intact. The DNA comes from many species, but no matter. They can be identified by the sequences recovered. Students are valuable in this effort and can quickly end up with enough DNA in a plastic tube to work for a long time. Soon they learn to sequence DNA and analyze it. Finding something useful tends to concentrate their minds.
The Keanumycins (after Keanu Reeves), came from Pseudomonas bacteria living in the fluid of a Dictyostelium fruiting body, descendants of the one Dr. Strassmann found in Virginia. That niche was composed of Dictyostelium amoebae that had transformed into a fruiting body that had a droplet of a few microliters at the top of a stalk. The droplet (our niche) had about 80,000 tough spores. It also has Pseudomonas bacteria called QS1027, that floats outside the spores. Sometimes a nematode crawls up the stalk and writhes in the droplet of our little community, making it shake. Shaking fruiting bodies with worms in them are a little freaky the first time you see them. Victor Zaydfudim, a high school student in our lab noticed them 20 years ago.
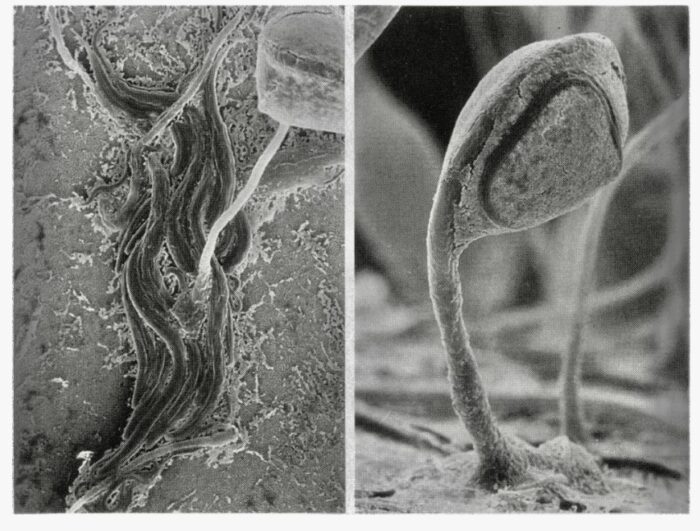
On the left, adult nematodes crawl around a Dictyostelium stalk. On the right, a resistant form of the nematode, called a dauerlarva, has crawled into the liquid droplet of our fruiting body, or niche, where antibiotic secreting pseudomonas live.
What do the Keanunmycins do? There are three, plus several others that were detected earlier. They do not kill bacteria. Rather, they punch holes in cell membranes of amoebae and fungi, which can be dangerous pathogens. Keanumycin A is a complex ring molecule with a two variants. One amoeba of Dictyostelium can eat 300 pseudomonas bacteria in an hour, but not when the bacteria make keanumycin and or a second molecule called jessinipeptin. These are lead natural product compounds for a new class of antibiotics.
Keanumycin A kills Dictyostelium at very low concentration, which is expected from its derivation, but it also kills several pathogenic Acanthamoeba species. The drug resistant yeast strain Candida auris, which can kill humans, is also controlled by keanumycin in vitro. (We are a long way from injecting these drugs).
The most important effects of keanumycin may be in agriculture because it kills Botrytis cinerea and other phytopathogens, Botrytis blight is a serious pest of greenhouse crops and vineyards. Pierre Stallforth and his colleagues have adopted the Hydrangea plant as their model organism. Botrytis infects hundreds of plants, so they chose one. I wonder if these or other natural products will help control diseases of our Ash, Beech, Chestnut and other trees.